The Jiggins Lab Webpage
 |
 |
 |
 |
 |
 |
 |
 |
Research
Genetic variation in susceptibility to parasites
In most organisms there is a lot of genetic variation in the susceptibility of individuals to parasitism. Yet this appears paradoxical. Being resistant is clearly advantageous, so why hasn't natural selection eliminated the susceptible alleles from the population? We are identifying the genes and mutations that cause this variation in wild Drosophila populations. This will allow us to understand both the molecular causes of this variation, and ultimately the evolutionary reasons why natural selection hasn't eliminated susceptibility alleles from the population. To tackle this question we have been using linkage mapping and genome-wide association studies (GWAS) in investigate the genetic basis of resistance viruses and parasitoid wasps in Drosophila. In many cases, we have found that there is a remarkably large amount of genetic variation in resistance, much of which is explained by a small number of major-effect resistance polymorphisms (e.g. Figure 1).
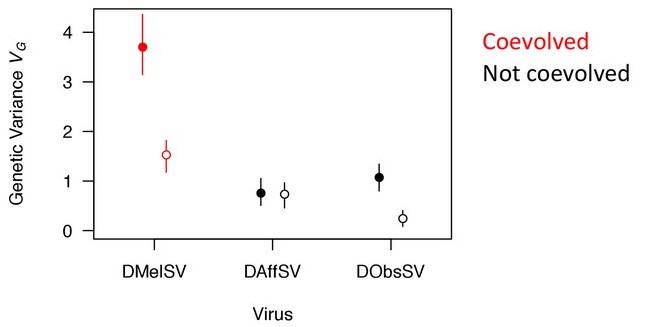
Figure 1. There is more genetic variation in susceptibility to viruses that have naturally coevolved with Drosophila (closed symbols). This effect largely disappears when a handful of major-effect resistance genes are accounted for (open symbols).
The evolution of immune systems
Host-parasite interactions are commonly associated with rapid evolution. This occurs because hosts are continually evolving novel defences, while parasites evolve to evade those defences. We have been using physiological and genetic approaches to understand how the immune system changes as hosts evolve resistance to infection. For example, we recently found that Drosophila evolves resistance to parasitoid wasps by constitutively activating its cellular immune response (Figure 2).
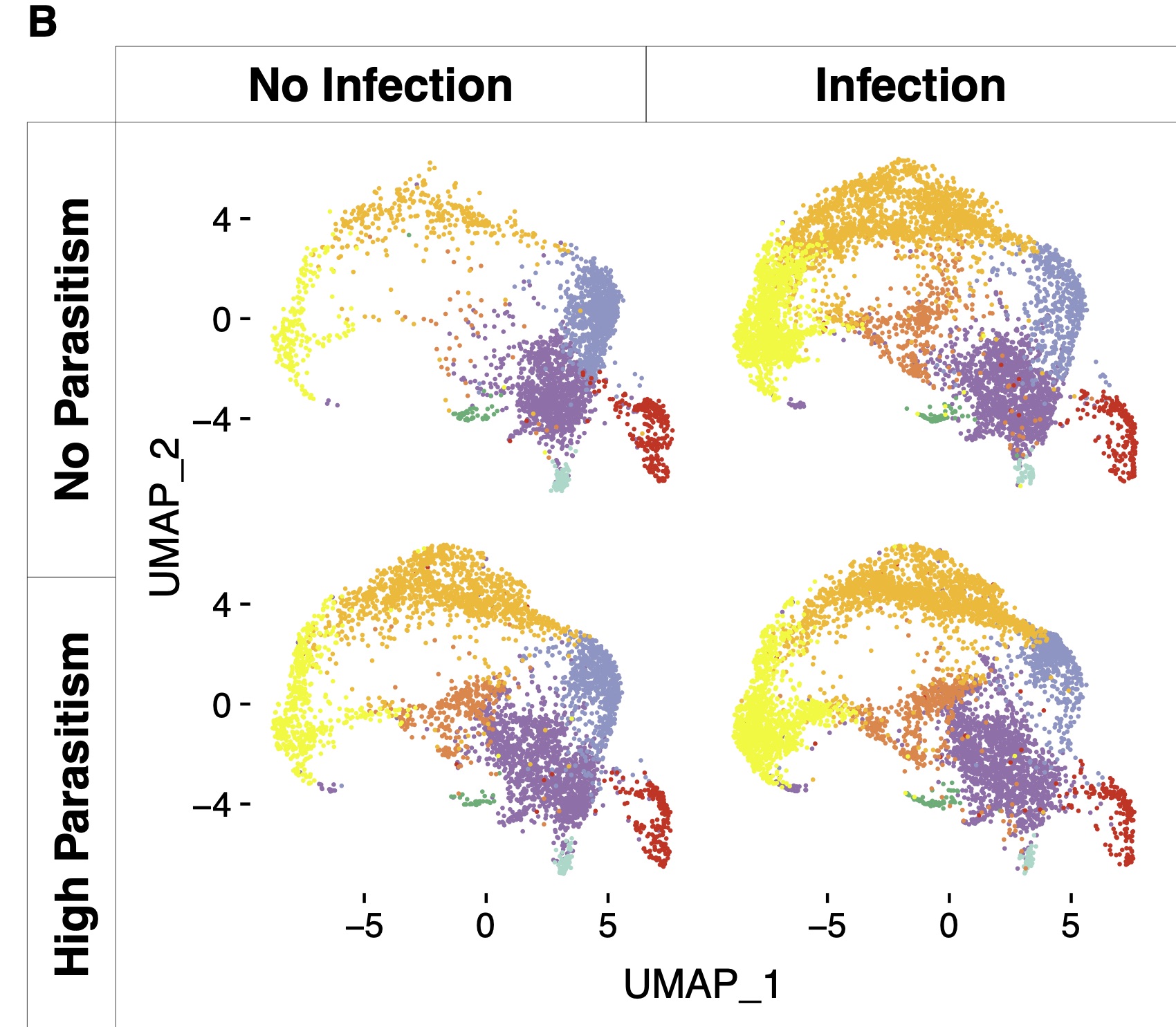
Figure 2. Single cell RNA sequencing of Drosophila blood cells. Infection results in an induced increase in specialised immune cell types (infection vs no infection), while this response is genetically hard-wired in flies that had evolved resistance to infection (parasitism vs no parasitism).
Mosquitoes
Insect vectored diseases are of considerable medical importance, and we are working one of the most important vector species, the mosquito Aedes aegypti. Currently we are collaborating with a public health program that aims to block dengue virus transmission by releasing mosquitoes infected with the bacterial symbiont Wolbachia, and using genetic approaches to understand the spread of the mosquitoes out of Africa. We have excellent insectary facilities for infection and survival assays.

Figure 4. Aedes aegypti (Source: CDC).
Myxoma virus
The introduction of the myxoma virus into populations of rabbits in Europe and Australia caused massive mortality, and imposed strong selection for increased resistance in the rabbit population and reduced virulence in the virus population. We are investigating the genetic basis of the evolutionary changes that occurred in the host and parasite populations since the virus was released.

Figure 5. European Rabbits (Source: Wikimedia Commons).